The previous blog post might have been a bit too dense to start with, so I’ll try something a bit lighter this time like importing RDF data into Neo4j. It assumes, however, a certain degree of familiarity with both RDF and graph databases.
There are a number of RDF datasets out there that you may be aware of and you may have asked yourself at some point: “if RDF is a graph, then it should be easy to load it into a graph database like Neo4j, right?”. Well, the RDF model and the property graph model (implemented by Neo4j) are both graph models but with some important differences that I wont go over in this post. What I’ll do though, is describe one possible way of migrating data from an RDF graph into Neo4j’s property graph database.
I’ve also implemented this approach as a Neo4j stored procedure, so if you’re less interested in the concept and just want to see how to use the procedure you can go straight to the last section. Give it a try and share your experience, please.
The mapping
The first thing to do is plan a way to map both models. Here is my proposal.
:Resource and have a property uri with the resource’s URI.(S,P,O) => (:Resource {uri:S})...(S,P,O) && isLiteral(O) => (:Resource {uri:S, P:O})(S,P,O) && !isLiteral(O) => (:Resource {uri:S})-[:P]->(:Resource {uri:O})ex:index.html dc:creator exstaff:85740 .
ex:index.html exterms:creation-date "August 16, 1999" .
ex:index.html dc:language "en" .ex stands for http://www.example.org/, exterms stands for http://www.example.org/terms/, exstaff stands for http://www.example.org/staffid/ and dc stands for http://purl.org/dc/elements/1.1/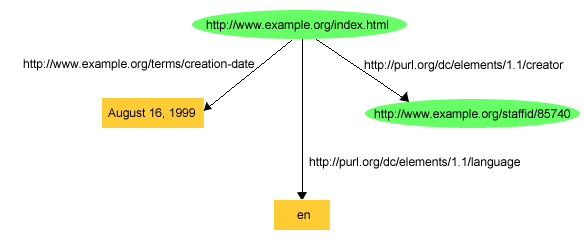
(:Resource { uri:"ex:index.html"})-[:`dc:creator`]->(:Resource { uri:"exstaff:85740"})(:Resource { uri:"ex:index.html", `exterms:creation-date`: "August 16, 1999"})
(:Resource { uri:"ex:index.html", `dc:language`: "dc"})
Categories
rdf:type). So let’s add a new rule to deal with this case.rdf:type statements are mapped to categories in Neo4j.(Something ,rdf:type, Category)=>(:Category {uri:Something})
:Person which makes a lot more sense and there is no semantic loss.The naming of things
Datatypes in RDF literals
Literals can have data types associated in RDF by by pairing a string with a URI that identifies a particular XSD datatype.
exstaff:85740 exterms:age "27"^^xsd:integer .As part of the import process you may want to map the XSD datatype used in a triple to one of Neo4j’s datatypes. If datatypes are not explicitly declared in your RDF data you can always just load all literals as Strings and then cast them if needed at query time or through some batch post-import processing.
Blank nodes
The building block of the RDF model is the triple and this implies an atomic decomposition of your data in individual statements. However -and I quote here the W3C’s RDF Primer again- most real-world data involves structures that are more complicated than that and the way to model structured information is by linking the different components to an aggregator resource. These aggregator resources may never need to be referred to directly, and hence may not require universal identifiers (URIs). Blank nodes are the artefacts in RDF that fulfil this requirement of representing anonymous resources. Triple stores will give them some sort of graph store local unique ID for the purposes of keeping unicity and avoiding clashes.
Our RDF importer will label blank nodes as BNode, and resources identified with URIs as URI, however, it’s important to keep in mind that if you bring data into Neo4j from multiple RDF graphs, identifiers of blank nodes are not guaranteed to be unique and unexpected clashes may occur so extra controls may be required.
The importRDF stored procedure
UPDATE [Feb-2021] The stored procedure described here in its initial form, evolved into a toolkit called Neosemantics (n10s) that includes a number of features that make it possible to work with RDF in Neo4j. The syntax described below has changed since, and it would be impractical to try and keep this post up to date. For the latest on the implementation have a look at the manual or check out the github repository for the source.
As I mentioned at the beginning of the post, I’ve implemented these ideas in a plugin for Neo4j called Neosemantics (n10s), which includes a procedure called importRDF. The usage is pretty simple. It takes four arguments as input.
- The url of the RDF data to import.
- The type of serialization used. The most frequent serializations for RDF are JSON-LD, Turtle, RDF/XML, N-Triples and TriG. There are a couple more but these are the ones accepted by the stored proc for now.
- A boolean indicating whether we want the names of labels, properties and relationships shortened as described in the “naming of things” section.
- The periodicity of the commits. Number of triples ingested after which a commit is run.
CALL semantics.importRDF("file:///Users/jbarrasa/Downloads/opentox-example.turtle","Turtle", false, 500)
Will produce the following output:
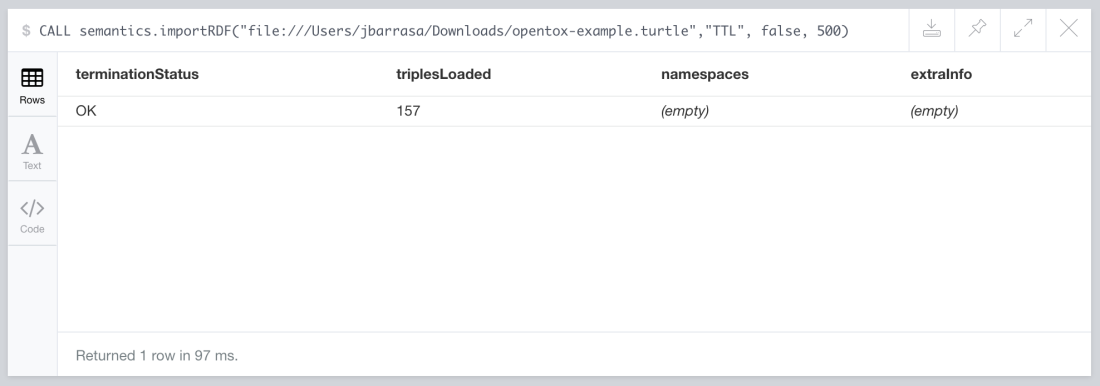
The URL can point at a local RDF file, like in the previous example or to one accessible via HTTP. The next example loads a public dataset with 3.5 million triples on food products, their ingredients, allergens, nutrition facts and much more from Open Food Facts.
CALL semantics.importRDF("http://fr.openfoodfacts.org/data/fr.openfoodfacts.org.products.rdf","RDF/XML", true, 25000)
On my laptop the whole import took just over 4 minutes to produce this output.
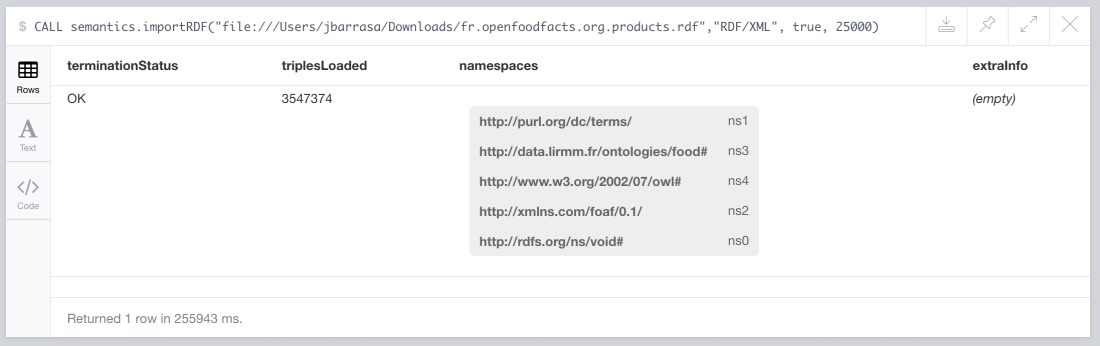
When shortening of names is selected, the list of prefix being used is included in the import summary. If you want to give it a try don’t forget to create the following indexes beforehand, otherwise the stored procedure will abort the import and will remind you:
CREATE INDEX ON :Resource(uri) CREATE INDEX ON :URI(uri) CREATE INDEX ON :BNode(uri) CREATE INDEX ON :Class(uri)
Once imported, I can find straight away what’s the set of shared ingredients between your Kellogg’s Coco Pops cereals and a bag of pork pies that you can buy at your local Spar.
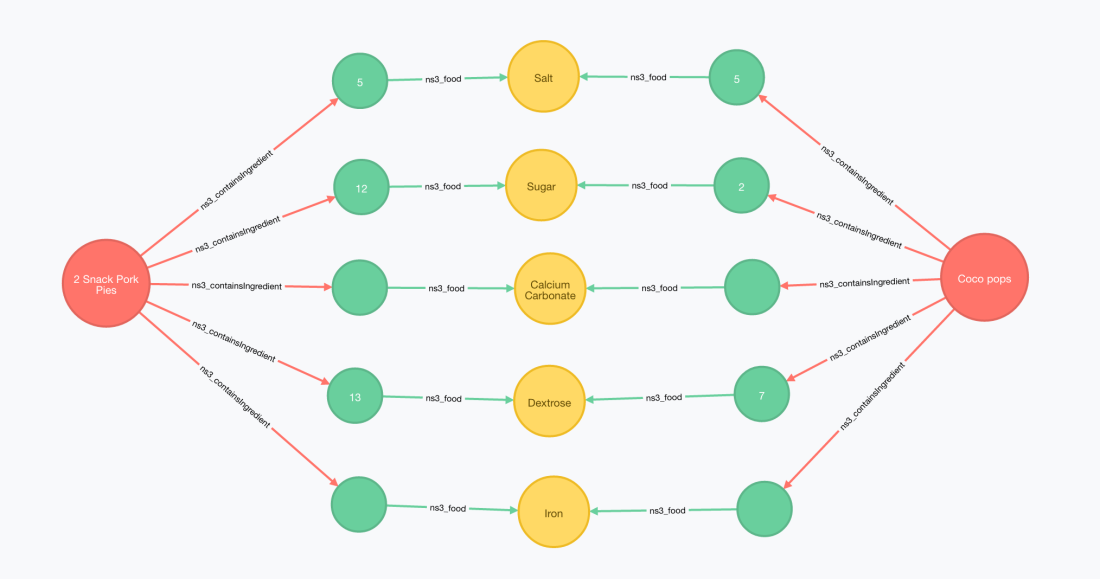
Below is the cypher query that produces these results. Notice how the urls have been shortened but unicity of names is preserved by prefixing them with a namespace prefix.
MATCH (prod1:Resource { uri: 'http://world-fr.openfoodfacts.org/produit/9310055537194/coco-pops-kellogg-s'})
MATCH (prod2:ns3_FoodProduct { ns3_name : '2 Snack Pork Pies'})
MATCH (prod1)-[:ns3_containsIngredient]->(x1)-[:ns3_food]->(sharedIngredient)<-[:ns3_food]-(x2)<-[:ns3_containsIngredient]-(prod2)
RETURN prod1, prod2, x1, x2, sharedIngredient
I’ve intentionally written the two MATCH blocks for the two products in different ways, one identifying the product by its unique identifier (URI) and the other combining the category and the name.
A couple of open points
There are a couple of thing that I have not explored in this post and that the current implementation of the RDF importer does not deal with.
Mutltivalued properties
The current implementation does not deal with multivalued properties, although an obvious implementation could be to use arrays of values for this.
And the metadata?
This works great for instance data, but there is a little detail to take into account: An RDF graph can contain metadata statements. This means that you can find in the same graph (JB, rdf:type, Person) and (Person, rdf:type, owl:Class) and even (rdf:type, rdf:type, refs:Property). The post on Building a semantic graph in Neo4j gives some ideas on how to deal with RDF metadata but this is a very interesting topic and I’ll be coming back to it in future posts.
Conclusions
Migrating data from an RDF graph into a property graph like the one implemented by Neo4j can be done in a generic and relatively straightforward way as we’ve seen. This is interesting because it gives an automated way of importing your existing RDF graphs (regardless of your serialization: JSON-LD, RDF/XML, Turtle, etc.) into Neo4j without loss of its graph nature and without having to go through any intermediate flattening step.
The import process being totally generic results in a graph in Neo4j that of course inherits the modelling limitations of RDF like the lack of support for attributes on relationships so you will probably want to enrich / fix your raw graph once it’s been loaded in Neo4j. Both potential improvements to the import process and post-import graph processing will be discussed in future posts. Watch this space.
Hello Jesús,
i’m new to the Neo4J world and interested to import rdf N-Triples data to Neo4J. So I got to your blog post. At the moment i am a little bit stucked, because i don’t know how to use your procedure. This are the steps that i’ve done:
1. Downloaded your source code
2. Generate a jar file with ‘mvn package’
3. Put the generated jar in the plugin folder of Neo4J
4. Called the procedure over the web interface
But calling the procedure generates the error:
‘There is no procedure with the name `semantics.importRDF` registered for this database instance. Please ensure you’ve spelled the procedure name correctly and that the procedure is properly deployed.’.
Can you give me some suggestions how to solve it? Maybe APOC is a necessary dependency?
Best regards
Timo
LikeLiked by 1 person
Hi Timo, great to hear that you’re giving the loader a try! You seem to have followed the right steps but let me ask you a couple of questions.
1. Did you restart the Neo4j server after copying the jar to the plugins folder?
2. Did you copy across also the dependent jars? APOC is not a required jar in the current version but there are a number of required third party jars that you’ll find in the pom.xml that need to be copied to the plugins directory as well.
3. Finally, if you’ve checked the previous two and it still does not work I’d recommend opening an issue in github (look at this simliar one https://github.com/jbarrasa/neosemantics/issues/1 ). At this point maybe the version of Neo4j you’re working with and the server logs would be useful.
Hope this helps. Let me know how it goes.
JB.
LikeLike
Hey,
after adding the other jars in the plugin folder as well, it’s working fine.
Thanks a lot! 🙂
LikeLiked by 1 person
That’s great!
I’d love to hear more about your experience. Also feedback/contributions to the loader are more than welcome.
JB.
LikeLike
Hello,
I have some trouble with your procedure…
In Neo4j, callling the procedure returns always a KO termination status with the same extra info:
“At least one of the required indexes was not found [ :Resource(uri), :URI(uri), :BNode(uri), :Class(uri) ]”
Even your example
CALL semantics.importRDF(“http://fr.openfoodfacts.org/data/fr.openfoodfacts.org.products.rdf”,”RDF/XML”,false,5000)
returns the same issue.
Any idea?
Thank you for your help,
Thomas
LikeLike
Hi Thomas,
I did add this stopper to avoid kicking of the RDF load if the indexes were not present because without indexes it can be really slow on large RDF imports.
All you have to do is create the indexes as described in the post. You can do this either from the browser or from the shell by running the following instructions:
CREATE INDEX ON :Resource(uri)
CREATE INDEX ON :URI(uri)
CREATE INDEX ON :BNode(uri)
CREATE INDEX ON :Class(uri)
You can check that the indexes have been created by running :SCHEMA on your Neo4j browser.
Once the indexes are present the importRDF procedure should work nicely.
Let me know if this solves the problem.
Cheers,
JB.
LikeLike
Hello Jesús,
Nice, it works great!
Very good work, thank you very much for your help.
Thomas
LikeLike
Great to hear Thomas. I’d be interested to hear/read about your experience.
Also watch this space for more on integration Neo4j – RDF.
JB.
LikeLike
Hello there,
Unfortunately I still run into trouble getting it to work.
Let me recap the steps I took in case I did anything wrong (neo4j version 3.0.6):
1. downloaded the entire neosemantics-folder into maven/bin directory, running mvn package shade:shade
2. copied the target output unto neo4j/plugins save for original-neosemantics-1.0-SNAPSHOT.jar, which causes a crash at neo4j startup
3. CALL semantics.importRDF(“http://fr.openfoodfacts.org/data/fr.openfoodfacts.org.products.rdf”,”RDF/XML”, true, 25000) causes following error: At least one of the required indexes was not found [ :Resource(uri), :URI(uri), :BNode(uri), :Class(uri) ]
4. copied the JARs from “alternate location” directly into neo4j/plugins folder, restart server, neo4j crashes altogether.
I uploaded the error/warning portion from my log bc it would explode this comment section (http://incinerator.lima-city.de/errors.txt) I don’t understand the error because all the mentioned JARs are in the plugins folder…
Maybe you know what could be the matter?
Thanks,
Christian
LikeLike
For goodness sake, why can’t I read instructions properly? It was the creating Index issue, which was even pointed out in the original blog article AND was mentioned in this very comment section. Sometimes you can’t see the forest for the trees I guess.
Seems to work now, sorry for posting the question
Christian
LikeLike
No problem Christian, great to hear you found the solution and thanks for givint it a try.
Feedback is most welcome.
Cheers,
JB.
LikeLike
Hi Jesús,
Thanks for the detailed blogs and code that you have made availble to us “online community”. It had helped me a lot to get a running start on combining the two worlds RDF and Neo4j {graphs} with each other.
I have a conundrum with a real-life use-case and would like to know if you can get in touch with me. I’m intressted in your view on few points…..
Kind regards, Black
LikeLike
Hi Black, great to hear you’ve found the materials useful.
Drop me a line at jesus at dot com and let’s have a chat.
Cheers,
JB.
LikeLike
Hi again Black, sorry just noticed the email address was not displayed as expected there is a neo4j missing between the at and the dot com.
Cheers,
JB.
LikeLike
Hello Jesús,
I really searched something like this. Thank you for providing it! I tried today your loader and it is working fine. The only problem is scalability. I’m trying to import an 100 million triple file and it takes very long. In fact I have the impression that number of triples imported per second is going down. I’m trying on a 60Gb RAM machine. Did you try on larger datasets? Have you some ideas what is happening?
Thank you
Dennis
LikeLike
Hi Dennis,
The degradation in write performance is expected. In Neo4j all connections (relationships) between nodes are materialised at write time as opposed to triple stores where they are computed via joins at query time. It’s a trade-off between write and read performance. What you’re getting with Neo4j is more expensive transactional data load but lightning speed traversals at read time.
Also Keep in mind that this approach is transactional which may or may not be the best approach for super large datasets. If your dataset is really massive you may want to try the non transactional import tool (https://neo4j.com/docs/operations-manual/current/import/)
The largest data set I’ve imported with my stored procedure is 107Million triples and it took 2h50min on my 16Gb laptop.
So I’d say it depends, if it’s a one off import it can be acceptable but if you want to import 100Mill triples every hour then probably you’ll need to find alternatives.
BTW, I’m currently working on the next post describing the experience of loading the larger dataset I mentioned before so watch this space.
Cheers,
JB.
LikeLike
Hello Jesús,
thank you for your answer, it is very helpful. I will see ahead for the next post!
Salut
Dennis
LikeLike
Hi Jesus,
thank you for your work that allows the Semantic Web community to use more easily neo4j technology. I wrote once in the post about importing RDF datasets. My problem was that I had problems importing a bigger RDF dataset in neo4j. Unfortunately I still have the problem. You pointed me to the neo4j-import utility. So I did the following. I parsed an RDF file and I created a dictionary for all the nodes using a quite big HashMap. Then I created two files:
reduced_dbpedia_nodes.csv:
id:ID,uri,:LABEL
7,”http://dbpedia.org/resource/Albedo”,Resource
reduced_dbpedia_relations.csv:
:START_ID,:END_ID,:TYPE
1,2,”http://www.w3.org/1999/02/22-rdf-syntax-ns#type”
I used neo4j-import for importing. I can import a part of DBpedia (and it is fine) but when I want to load also the yago classes (just a lot of classes) I get a strange error I reported here:
http://stackoverflow.com/questions/41909362/neo4j-import-fails
You said that you wanted to do a post for importing big datasets. Is this still the case? Can you help?
Merci
Dennis Diefenbach
LikeLike
Hi Dennis, I see you uncovered a bug in the import tool. Thanks and nice catch.
I thought I’d drop you a line to mention that one year later… yes, that’s how time flies and how other priorities take precedence 😦 I’m about to publish a worked example on a larger dataset.
I’m using the OpenPermID from Thomson Reuters.
Should be public in the next week or so.
Thanks for the patience 🙂
JB.
LikeLike
Thanks for your code.But when I running the code CALL semantics.importRDF(“http://fr.openfoodfacts.org/data/fr.openfoodfacts.org.products.rdf”,”RDF/XML”, { languageFilter: ‘fr’, commitSize: 5000 , nodeCacheSize: 250000}) the procedure returns always a KO termination status with the same extra info:”unqualified property element not allowed [line 2, column 14]”
LikeLiked by 1 person
Hi, yes. That was reported a few weeks ago.
Find the solution here https://github.com/jbarrasa/neosemantics/issues/31
Let me know if this solves the problem.
Enjoy!
JB.
LikeLike
Hello,
Thank you for the great article.
I have followed the procedure for loading the triples in Neo4j but i am getting the following error:
“Neo.ClientError.Statement.SyntaxError: Procedure call does not provide the required number of arguments: got 4 expected 3.”
This is with the example that you have provided:
CALL semantics.importRDF(“http://fr.openfoodfacts.org/data/fr.openfoodfacts.org.products.rdf”,”RDF/XML”, true, 25000)
Any help will be appreciated.
Thanks and regards,
Arun
LikeLike
Hi Arunava, the stored procedure has evolved since I published this blog post back in 2016.
Please have a look at the documentation for the current version of the importRDF procedure in GitHub.
Manual here: https://github.com/jbarrasa/neosemantics
Regards,
JB.
LikeLike
Hi, thanks for this wonderful work. But I have a small trouble while getting the names of the nodes. It shows URI in place of them, can you help me with this.
Thanks in advance
LikeLike
Hi PJ, thanks for your interest. When you say “it shows” you mean when you visualise the imported data in the Neo4j browser?
If that’s the case, here’s how to style the visualisation https://neo4j.com/developer/guide-neo4j-browser/
Cheers,
JB
LikeLike
Thank you for your immediate reply. What I mean in the above question is, I am unable to get the labels of the nodes. Only uri is seen for each node
LikeLike
Hi, Jesús!
Now I want to import RDF N-Triples to neo4j. But I have some difficulties. The following steps are all under Windows.
First, I downloaded the .jar file from github, and then I put it into the plugins folder of neo4j. And then I add “dbms.unmanaged_extension_classes=semantics.extension=/rdf” to conf/neo4j.conf. In the end, I restart the server and run “call dbms.procedures()”, but “semantics.*” didn’t appear in the list.
Now I have no idea what I can do. Can you give me some suggestions? Thank you!
LikeLike
Now I succeed by building the JARs from the source. Although I suffered some errors in the way, I solved them. Thank you for your wonderful work,again!
LikeLike
Hi June, thanks for using neosemantics! It’s interesting that you had to rebuild from source. I’d like to understand what the cause of the problem was and similarly what sort of errors you found during the build.
That said, probably this kind of conversation makes a lot more sense in the issues section of the github repo (https://github.com/jbarrasa/neosemantics/issues).
JB.
LikeLike
Hello Jesus,
For a while I was trying to find a straightforward way how to port RDF into neo4j, until I found your great post (and its sequels) – thank you for sharing all this! I went through all suggested steps: the .jar file loaded well into plugins, after catching a few errors of my own I went to executing the stored procedure (the latest version from your GitHub, with syntax chosen accordingly). I got stuck when trying to load into ne4j the following .ttl (or, in fact, any from the BBC repository):
CALL semantics.importRDF(“https://github.com/bbc/curriculum-data/blob/master/maths.ttl”,”Turtle”, { shortenUrls: false, typesToLabels: true, commitSize: 500 })
… the loader then terminated with KO and extraInfo “IRI included an unencoded space: ’32’ [line 7]”.
When I downloaded the file to desktop and tried to load the file into neo4j locally:
CALL semantics.importRDF(“file:///C:/…/maths.ttl”,”Turtle”, { shortenUrls: false, typesToLabels: true, commitSize: 500 })
… I get still KO, with a different extraInfo “Expected an RDF value here, found ‘=’ [line 58]”.
Then I tried to replicated literally – just with the latest version as per GitHub – your post example, with:
CALL semantics.importRDF(“http://fr.openfoodfacts.org/data/fr.openfoodfacts.org.products.rdf”,”RDF/XML”, { shortenUrls: false, typesToLabels: true, commitSize: 25000 })
… still KO, with extraInfo “unqualified property element not allowed [line 2, column 14]”.
Being new to RDF and neo4j I am sure I am doing some rookie mistake somewhere, but neither searching nor experimenting took me out of this – your kind hint would be much appreciated. Thank you!
LikeLike
Hi Vladislav, when loading the BBC data from GitHub you should use the raw version of the Turtle file, otherwise you’ll be trying to load an html page which is, of course, not valid RDF. You can see the page by clicking on the ‘Raw’ button on the top right of the source code in GitHub. This is the correct url to use in the stored proc:
CALL semantics.importRDF(“https://raw.githubusercontent.com/bbc/curriculum-data/master/maths.ttl”,”Turtle”, { shortenUrls: false, typesToLabels: true, commitSize: 500 })
Regarding the openfoodfacts, their datasets are broken. I suggested interested people to reach out to them to have it fixed. All the parser can do is detect a syntax error but the file should be fixed at source. Here’se the issue in GitHub where this was tracked: https://github.com/jbarrasa/neosemantics/issues/31#issuecomment-427877700
By the way, these kind of technical questions are best tracked in GitHub than here.
Good luck and let me know how things go.
JB.
LikeLike
Hi Jesus, thank you – it moved me a step! However, not yet till the very end: loading the file from the raw GitHub still ends with an error “”Expected an RDF value here, found ‘=’ [line 58]” (which is the same loader error as when I donwloaded the file to desktop and used the neo4j loader locally).
Next such question I will move to the GitHub, as you suggested.
Thank you!
LikeLike
what formats does this stored procedure support? My data is currently in compressed N-Triples format….
LikeLike
Supported serialisation formats are: Turtle, N-Triples, JSON-LD, TriG, RDF/XML
All described in the readme of the Github repo: https://github.com/jbarrasa/neosemantics
LikeLike
I was able to import one of my “Descriptors’, namely https://id.nlm.nih.gov/mesh/D002318.rdf, but I am not brave enough to import all the triples on my personal laptop at this hour. I will try tomorrow at work.
LikeLike
That’s great to hear. Looking forward to hearing how the large dataset import goes.
LikeLike
Hi Jesus,
I was trying to address the multivalued property issue by editing some of your code in the setProp function in the DirectStatementLoader class where you commented:
// we are overwriting multivalued properties.
// An array should be created. Check that all data types are compatible.
When I try to add an array to the map and run the RDFImportTest, I get a error:
org.neo4j.driver.v1.exceptions.ClientException: Failed to invoke procedure `semantics.importRDF`: Caused by: java.lang.IllegalArgumentException: [[Ljava.lang.ArrayList;] is not a supported property value
LikeLike
Hi Marc, yes, there is not an easy solution. Check out this discussion: https://github.com/jbarrasa/neosemantics/issues/18
Also, this conversation on code is better addressed as issues in the GitHub repo than here in the blog so let’s move there if that is ok.
LikeLike
Hi Jesus, fantastic post and super helpful for someone getting into neo4j like myself. Do you have a followup on how to get RDF metadata, as mention in the post? I’m trying to get the rdf:type of an instance but can’t figure out how. I have a query like:
MATCH (n:owl__NamedIndividual)-[*..3]->(m:owl__Class)
WHERE n.rdfs__label = ‘2017 Volkswagen GTI Sport’
RETURN n, m
LIMIT 25
And I’d like to get ‘m’ node back where the label would be ‘GTI’ given the following class heirarchy:
Automobile ➝ Volkswagen ➝ Compact ➝ GTI ➝ “2017 Volkswagen GTI Sport”
LikeLike
Hi Sammy, thanks. Happy to hear you’re finding it useful.
Answering your question, your query looks reasonable but it really depends on how you’ve carried out the data import and what’s in the dataset you’ve imported.
Statements of type `rdf:type` can be imported in two ways: (1)as LPG labels or (2) as separate nodes depending on the value of the `typesToLabels` parameter. I’m afraid you’ll have to share the `importRDF` call you’ve used and at least a part of the dataset if you want me to try it at my end. Is that possible?
RDF metadata is itself RDF so in principle there should be no difference. But again i should be able to respond more precisely with a dataset we can comment on.
One final comment, could you bring this conversation over to github issues (https://github.com/jbarrasa/neosemantics/issues), please? I think it’s a more adequate platform for technical discussions.
Thanks!
JB.
LikeLike
Sure thing, just posted the issue: https://github.com/jbarrasa/neosemantics/issues/65
Unrelated, I watched your presentation “How Semantic Is Your Graph?” from GraphConnect 2016, and was wondering if the demo was available for download somewhere? In the video, you had it running locally at /legis-graph/sem-demo.html
Thanks!
LikeLike
Hi Jesus,i have import owl file successfully,but in the graph,every node didn’t shows its lable but a uri(like uri: http://www.semanticweb.org/asus/ontologies/2019/4/untitled-ontology-10#map)
Because of this,i can’t know what every node means.
How to work it out?
LikeLike
In the link below you can find how to change the styling used by Neo4j browser. This way you can choose the properties you want displayed instead of the uri.
For technical questions on neosemantics please use GitHub issues instead of comments here. Thanks!
https://neo4j.com/developer/guide-neo4j-browser/#_styling_neo4j_browser_visualization
LikeLike
Hi Jesus,
Even I faced the same issue of indexes.
Can you please explain me the purpose of indexes?
Regards
Prashanti
LikeLike
Hi Prashanti, if you are referring to the index on :Resource(uri), it’s needed to accelerate the many lookups needed to link resources to one another using relationships. Before you run any RDF import the index needs to be created.
Hope this helps. Let me know if I misunderstood your question.
JB
LikeLike
Yes. Thank you for your prompt response.
LikeLike
Hello Jesus,
I have my own rdf file which I am trying to load into Neo4j using Call semantics.importRDF(“file:///C:/Users/prash/OneDrive/Documents/Prashanti/RdfFile/myrdf.rdf”, “Turtle”, {})
But I am facing “Expected ‘.’, found ‘<' [line 1]" error. Any idea what needs to be fixed?
LikeLike
It looks like it’s not a Turtle file but an RDF/XML. If that’s the case you’ll have to use:
semantics.importRDF(“file:///C:/…/myrdf.rdf”, “RDF/XML”, {})
LikeLike
No, it doesn’t work. Let me know if you want to take a look at my rdf file. I can email you.
LikeLike
Trying to import from local windows machine with below command but getting following error.
file Sample_Payload_1.json in import folder on local windows machine
//import jason-LD files in neo4j using neosematics plug in
call semantics.importRDF(“file:///Sample_Payload_1.json”, “JSON-LD”,{ shortenUrls: true, typesToLabels: true, commitSize: 9000 });
terminationStatus: KO
extraInfo: “\Sample_Payload_1.json (The system cannot find the file specified)”
thoughts? and ideas to resolve the issue?
LikeLike
Please, for these kind of questions go to github issues (https://github.com/neo4j-labs/neosemantics/issues).
There is actually a comment on windows path expression. See here (https://github.com/neo4j-labs/neosemantics/issues/62#issuecomment-444133096).
Hope this helps.
JB.
LikeLike
Are blank nodes still being labeled as BNode? I imported the STATO ontology using CALL semantics.importRDF(‘https://raw.githubusercontent.com/ISA-tools/stato/dev/releases/latest_release/stato.owl’, ‘RDF/XML’). But no node is labeled as BNode. There’re many blank nodes because of owl:restriction. What’s intersting is there’re also blank nodes of type owl:Class (labled as owl__Class after import).
I’m asking because I want to find a way to exclude the blank nodes in a search. Right now I’m using
MATCH (n:owl__Class) where exists(n.rdfs__label) RETURN n
But I think there must be a better way to do this.
LikeLike
Could you please open an issue in GitHub or a question in the community site? This way the whole community will benefit from the answer. Please do and I’ll take it from there.
Here are the links:
https://github.com/neo4j-labs/neosemantics/issues
https://community.neo4j.com/c/integrations/linked-data-rdf-ontology
Thank you!
JB
LikeLike
Hi
Any idea for be able to go over the limitations of RDF like the lack of support for attributes on relationships ?
I convert al my data to turtle how can i add to the relation attributes and insert them to neo4j?
LikeLike
The short answer is RDF*
You’ll have it in the next release of Neosemantics.
Watch this space.
LikeLike
Hi Jesús,
I am just getting started with exploring rdf data and neo4j in general using neosemantics. When I try to import the rdf dataset, I get an error with extraInfo stating “Unexpected character U+7C at index 52:..”. Is this related to the coding of the data and how can I fix it?
LikeLike
Hi!
Looks like it’s an encoding issue that is making the RDF parser choke.
For bugs, I’d suggest you raise an issue in GitHub (https://github.com/neo4j-labs/neosemantics/issues) and add details so we can reproduce.
And for tech questions probably the best is the RDF channel in the community site (https://community.neo4j.com/c/integrations/linked-data-rdf-ontology)
Cheers,
JB.
LikeLiked by 1 person
Thanks for the quick reply!
LikeLiked by 1 person
CALL semantics.importRDF(“file:///D:/All_Script/Protege/neo4j.turtle”,”Turtle”, true, 500),wrong
Procedure call provides too many arguments: got 4 expected no more than 3.
Procedure semantics.importRDF has signature: semantics.importRDF(url :: STRING?, format :: STRING?, params = {} :: MAP?) :: terminationStatus :: STRING?, triplesLoaded :: INTEGER?, triplesParsed :: INTEGER?, namespaces :: MAP?, extraInfo :: STRING?, configSummary :: MAP?
meaning that it expects at least 2 arguments of types STRING?, STRING?
Description: Imports RDF from an url (file or http) and stores it in Neo4j as a property graph. Requires and index on :Resource(uri) (line 1, column 1 (offset: 0))
“CALL semantics.importRDF(“file:///D:/All_Script/Protege/neo4j.turtle”,”Turtle”, true, 500)”
^
why???
LikeLike
There’s a comment highlighted in red indicating that the code in the post is no longer up to date and that you should go to the manual (also linked in the comment) for the most up to date reference on how to use it.
Also for technical questions like this one probably github or the neo4j community site are certainly better channels than this blog.
Loooking forward to hearing from you over there. Enjoy!
JB
LikeLike